The functional landscape of proteins
The functional landscape of a protein is the portion of its free energy landscape that corrresponds to the native state. The functional motions of the protein (catalysis, ligand binding, allosteric regulation) occur on this landscape which has a complex topology.
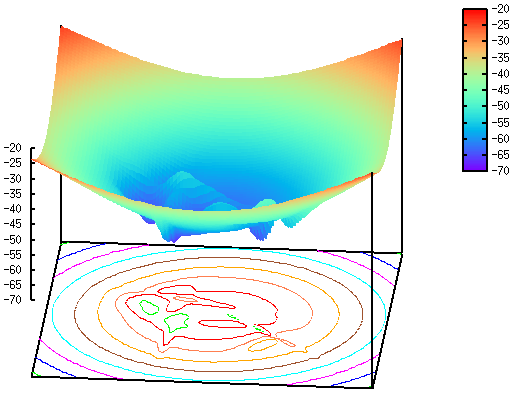
We plan to reveal the general properties (hierarchical nature, the type of dynamics, etc.) of this landscape on a structurally representative subset of proteins, focusing on proteins studied experimentally in our Institute. We plan to determine the functional dynamics of each protein by simulations of models representing various levels of coarse-graining, and a range of analysis methods. The functional landscape is particularly important for the understanding of allosteric mechanisms and allosteric signal transmission pathways. To this end, we construct in silico structural models of protein complexes, and describe communication pathways by network models.
Coupled folding and binding of homodimers
Collaborator: Dániel Györffy (PhD student)
Intrinsically disordered proteins are often involved in regulatory and signal transduction processes, and exert their function by binding to other proteins. This binding typically results in a disorder-to-order transition whereby the protein folds. In many cases, this process can be described as a disordered chain getting ordered on an ordered template. However, two disordered chains may also bind each other, forming an ordered homodimer. The mechanism of such ordering is an intriguing problem
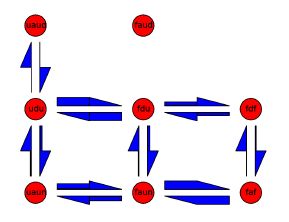
We use simplified protein models, including hydrophobic-polar lattice models, to elucidate the mechanisms of coupled binding and folding in homodimeric proteins. We describe the kinetics of the system by Markov state models (see figure above), determining reaction rates under various conditions.